New Larger Than Life Imaging of Model Worm Digs Up Finer Details

Despite being such a tiny earth dweller, the roundworm species Caenorhabditis elegans (C. elegans) has become one of the biggest workhorses in the lab for biological researchers. Due in part to the organism’s transparent skin and compact size — just about the size of a comma at 1 mm in length — it’s the only animal to have successfully had a complete mapping of its connectome, or its neural circuitry comprised of 302 neurons and their 7,000 synaptic connections.
Boasting this comprehensive neural map has made the modest worm a popular model for exploring fundamental principles that apply to the nervous systems of other animals, as well as many other questions. However, even today’s most powerful imaging technologies have lacked the capability to label neuronal connections and map the entire volume of the nervous system at once, making neuronal connections of the whole nervous system still hard to investigate.
Now, researchers have opened up greater possibilities for the study of C. elegans — unveiling a new imaging approach for capturing fine biological details of the nematode by physically blowing the sample up in size, rather than increasing magnification power of microscopes to investigate them.
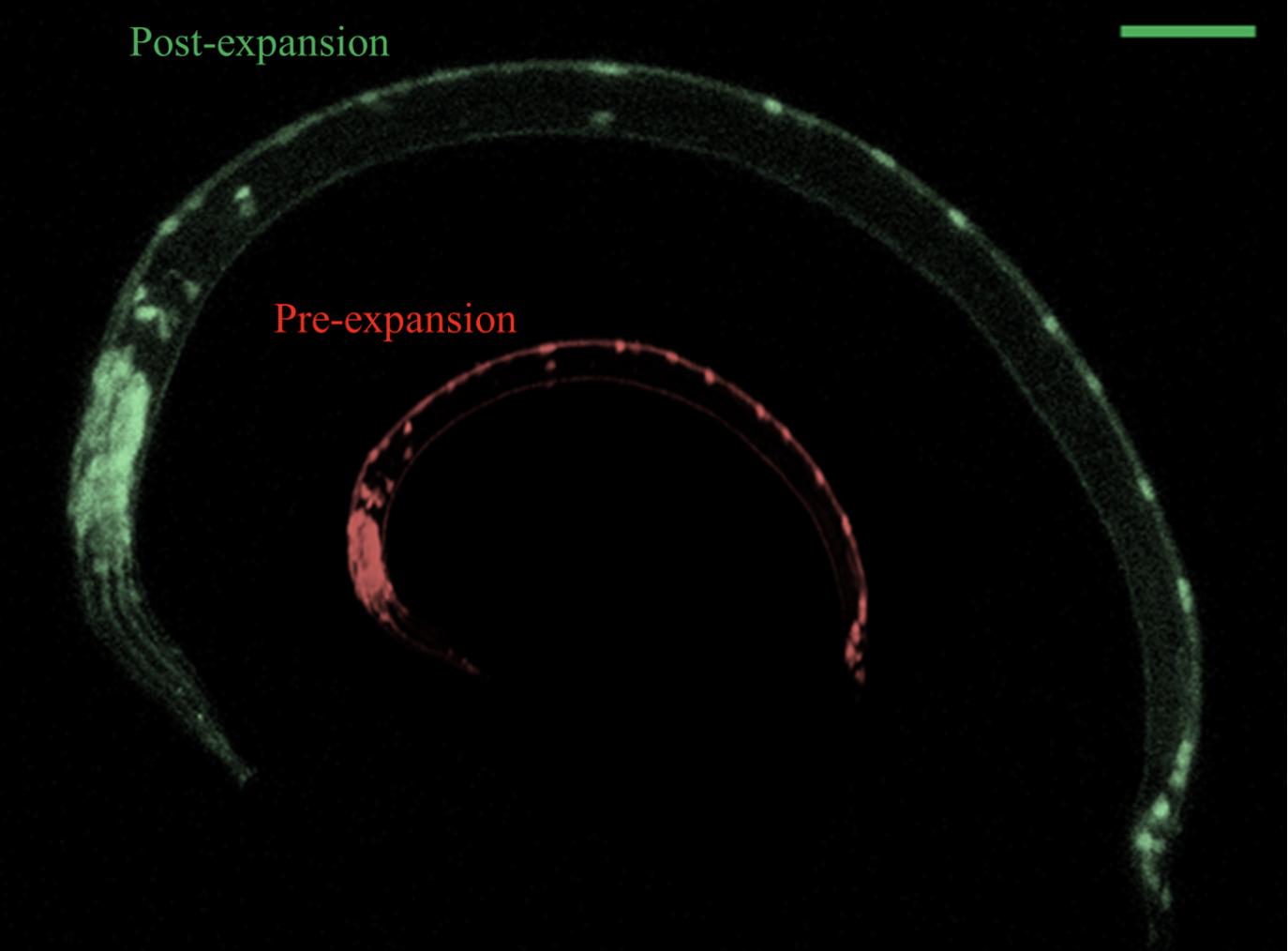
In findings reported in the journal eLife, NJIT, MIT and Stanford researchers have demonstrated their customized application of an up-and-coming imaging technique called expansion microscopy (ExM), which the team used to expand preserved samples of the worm by fivefold — up to 5 mm in size — enabling them to simultaneously map details from fluorescent proteins and RNA to DNA location and anatomical structures at resolutions of ~65–75 nanometers.
The study marks the first time the ExM technique — first demonstrated by MIT researchers led by Edward Boyden in 2015 — has been applied to image a whole animal. The team says the new approach could allow for a much more detailed study of the model organism that is accessible to a much broader range of laboratories equipped with conventional microscopes.
“The last time that all the synapses were mapped in a single animal was in 1986, and it took the researchers 15 years using electron microscopy to map a single nervous system. … The tracing of neurons and finding synapses were done manually on large photoprints,” said Gal Haspel, collaborating author of the paper and professor at NJIT’s Department of Biological Sciences. “The most important advancement with this new approach is that microscopic resolution can be improved by expanding the sample instead of improving the optics, which has given us a greater ability to see the nematode’s synaptic connections among neurons — only about 50 nanometers in size — with a standard confocal microscope.”
The collaborating teams developed a C. elegans-tailored version of expansion microscopy technology and named it expansion of C. elegans (ExCel) to overcome the specific challenge of physically expanding the nematode without damaging the biology contained within its highly rigid, multilayer protective exoskeleton. The expansion process generally involves embedding a preserved sample in an expandable polymer matrix that is roughly similar to the absorbing material found in disposable diapers. This forms a dense web of polymer threads spaced a few nanometers apart, serving as anchor points throughout the sample. Adding water causes the polymer matrix to expand. As the polymer threads swell apart from each other, biomolecules and labels within the sample are able to retain their spatial organization relative to one another.
“We had to dissolve the nematode exoskeleton without damaging the nervous tissue or the structures and proteins that we are interested in,” explained Zainab Tanvir, a Ph.D. student in the Haspel Laboratory. “We used digesting enzymes and chemical reactions that had to be optimized to an exact concentration and time to help us in expanding the tissue without losing integrity and relative positions inside it.”
Above: ExCel physically expands C. elegans. Near 2.5X expansion (scale bar: 50 micrometers). Credit — NJIT/MIT
With ExCel — capable of imaging fluorescent proteins after fivefold linear expansion — the team was able to locate the nematode’s synaptic connections. The team says that gaining the ability to florescent tag such proteins at such resolution could open new opportunities for more advanced studies of synaptic connections in other animals.
“These are known proteins that are in the synaptic locations, but we showed we could label them and find the synapses. … It previously wasn’t possible to label proteins and get enough resolution,” said Tanvir.
“What we would like to do now is to collect connectivity data from multiple animals and establish the variability of connectivity and its distribution in the nervous system,” added Haspel. “Variability is innately present at every level of biology and is what allows organisms to quickly adjust and react to their environment. So, we are interested in looking to see if variability is entirely stochastic in the nervous system or if there are other natural forces such as genetics that enable nervous systems to be variable.”
...
eLife 2020;9:e46249 DOI: 10.7554/eLife.46249